|
Using foot-and-mouth disease (FMD) as an example, this review describes new tools that can be used to detect and characterise livestock diseases. In recent years, molecular tests that can detect and characterise pathogens in a diverse range of sample types have revolutionised laboratory diagnostics. In addition to use in centralised laboratories, there are opportunities to locate diagnostic technologies close to the animals with suspected clinical signs. Work in this area has developed simple-to-use lateral-flow devices for the detection of FMD virus (FMDV), as well as new hardware platforms to allow molecular testing to be deployed into the field for use by non-specialists. Once FMDV has been detected, nucleotide sequencing is used to compare field strains with reference viruses. Transboundary movements of FMDV are routinely monitored using VP1 sequence data, while higher resolution transmission trees (at the farm-to-farm level) can be reconstructed using full-genome sequencing approaches. New technologies such as next-generation sequencing technologies are now being applied to dissect the viral sequence populations that exist within single samples. The driving force for the use of these technologies has largely been influenced by the priorities of developed countries with FMD-free (without vaccination) status. However, it is important to recognise that these approaches also show considerable promise for use in countries where FMD is endemic, although further modifications (such as sample archiving and strain and serotype characterisation) may be required to tailor these tests for use in these regions. Access to these new diagnostic and sequencing technologies in sub-Saharan Africa have the potential to provide novel insights into FMD epidemiology and will impact upon improved strategies for disease control.Effective control of infectious diseases is reliant upon accurate diagnosis of clinical cases using laboratory tests, together with an understanding of factors that impact upon the epidemiology of the infectious agent. A wide range of new diagnostic tools and nucleotide sequencing methods are used by international reference laboratories to detect and characterise the agents causing outbreaks of infectious diseases. In the past, high costs (initial capital expenses, as well as day-to-day maintenance and running costs) and complexity of the protocols used to perform some of these tests have limited the use of these methods in smaller laboratories. However, simpler and more cost-effective formats are now being developed that offer the prospect that these technologies will be even more widely deployed into laboratories particularly those in developing regions of the world such as sub-Saharan Africa.
This short review focuses on foot-and-mouth disease (FMD) and highlights new diagnostic approaches that can be used to detect and characterise pathogens causing livestock diseases. Foot-and-mouth disease is a trans-boundary viral disease that is endemic in sub-Saharan Africa, much of south Asia (Middle East, Indian sub-continent and Southeast Asia) and parts of South America. The causative agent, FMD virus (FMDV), is a picornavirus (genus: Aphthovirus) with a positive-sense RNA genome of approximately 8300 nucleotides in length. Foot-and-mouth disease virus exists as seven genetically discrete serotypes, five of which are currently endemic in sub-Saharan Africa. This virus causes an acute disease in cloven-hoofed animals (i.e. cattle, sheep, goats, pigs and buffalo) that is associated with the development of vesicles on epithelial surfaces of the mouth and feet. Foot-and-mouth disease virus infection also generates a transient viraemia in infected animals that typically lasts for approximately five days (Figure 1; Alexandersen et al. 2003). Tests that exploit these clinical windows in an infected animal form the basis of laboratory approaches currently used to diagnose FMD. These assays aim to detect FMDV in epithelium and fluid from vesicles, as well as in blood and swabs from mucosal surfaces (oral and nasal swabs). In addition, FMDV-specific antibody responses in exposed animals can be detected using serological assays; however these methods are not discussed in this review. In general terms, the virological assays utilise three different strategies: propagation of FMDV, detection of viral antigenic proteins, or use of molecular assays to amplify specific RNA sequences. The earliest versions of these tests exploited approaches that were largely used to propagate FMDV for vaccine production (Brown 2003). Although initially developed during the 1920s and used up until the 1950s, the obvious disadvantages of these systems placed limitations on their use for wide scale FMD diagnostics. Indeed, these systems were superseded by improved highly sensitive in-vitro cell cultures used to propagate FMDV, such as primary bovine thyroid cells (Snowdon 1966), permanent pig kidney cultures (IB-RS-2; De Castro 1964) and more recently a goat tongue cell line (Brehm et al. 2009). Virus isolation approaches can be highly sensitive (depending upon the cell culture system used) although it can be slow taking between one and four days to generate a result. Propagation of FMDV in cell cultures is still widely considered to be the ‘gold-standard’ test for FMD diagnosis. The development and establishment of cell culture systems has been paralleled by improvements to immunological (enzyme-linked immunosorbent assays: ELISA) assays designed to detect FMDV antigen using polyclonal antisera (Ferris et al. 1988) or characterised monoclonal antibody reagents (Ferris et al. 2011). Capture ELISAs are more rapid than virus isolation, but they have lower analytical sensitivity and are also inappropriate for use with certain sample types such as blood, milk and swabs. More recently, lateral-flow devices (LFDs, also referred to as immuno-chromatographic strip tests) have been developed for the detection of FMD viral antigen. These simple-to-use and rapid tests utilise FMDV specific antibody reagents (normally monoclonal antibodies) in a format similar to the sandwich capture ELISA used for laboratory diagnosis. Positive test signal is generated by the diffusion of coloured, antibody-coated latex beads or colloidal gold particles through a membrane towards an immobilising band of trapping antibody. An LFD has been developed for the detection of all seven FMDV serotypes which uses a pan-serotypic monoclonal antibody (Ferris et al. 2009). In addition, sample preparation in field conditions can be achieved using simple disposable tissue homogenizers for preparing epithelial suspensions. In terms of diagnostic sensitivity and specificity, the overall performance of this LFD is similar to laboratory-based antigen ELISA, although the diagnostic sensitivity of the current test is lower for SAT 2 field strains (Ferris et al. 2009). This LFD and a simple homogenising kit is now commercially available (via Boehringer Ingelheim Vetmedica Svanova, Uppsala, Sweden). Data from the field illustrates the potential for the LFD to be used in locations close to animals to provide rapid support to veterinarians in their clinical assessment of suspected FMD cases. The simplicity and stability of the LFD may be important features for FMD diagnosis in sub-Saharan African countries. In particular, ability to rapidly recognise FMDV in clinical material may improve the selection of diagnostic samples that are shipped to reference laboratories for subsequent isolation and/or strain characterisation. In addition, recent results have also indicated that it is possible to recover RNA from FMDV positive LFDs that can be used in rRT-PCR assays and sequencing studies. Further LFD assays are now in development and future formats, such as the SAT 2 device that has recently been evaluated (Ferris et al. 2010), may also allow rapid serotyping of field strains.
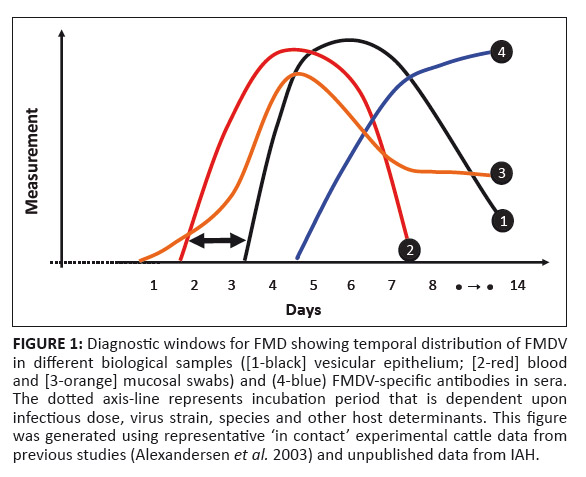 |
FIGURE 1: Diagnostic windows for FMD showing temporal distribution of FMDV
in different biological samples ([1-black] vesicular epithelium; [2-red] blood
and [3-orange] mucosal swabs) and (4-blue) FMDV-specific antibodies in sera.
The dotted axis-line represents incubation period that is dependent upon
infectious dose, virus strain, species and other host determinants. This figure
was generated using representative ‘in contact’ experimental cattle data from
previous studies (Alexandersen et al. 2003) and unpublished data from IAH.
|
|
|
Polymerase chain reaction technologies for foot-and-mouth disease virus detection
|
|
Amplification of specific nucleic acid sequences using reverse transcription polymerase chain reaction (RT-PCR) is now widely used for the laboratory detection of FMDV. These molecular assays are suitable for the diverse range of different samples that might be submitted for laboratory investigation (tissues, blood, swabs, oesophageal or pharyngeal (OP) scrapings, faecal samples and milk). Over the past 15 years, improvements have been made to RT-PCR protocols used for the detection of FMDV. Initially, assays that targeted conserved regions of the genome (3D: Meyer et al. 1991; Rodrıguez et al. 1994 and 5’ untranslated region [5’ UTR], Reid et al. 2000) utilised agarose gel electrophoresis for the detection of amplified products. However, these labour intensive procedures have a high risk of generating false positives due to carry-over of chain reaction (PCR) amplicons and are therefore not generally considered ideal for routine testing of large numbers of samples. Real-time RT-PCR (rRT-PCR) assays have now largely replaced agarose gel based assay formats. These more rapid fluorescence-based approaches are highly sensitive enabling simultaneous amplification and quantification of FMDV specific nucleic acid sequences. In addition to enhanced sensitivity, the benefits of these closed-tube rRT-PCR assays over conventional endpoint detection methods include a reduced risk of cross-contamination, their large dynamic range, an ability to be scaled up for high-throughput applications and the potential for accurate target quantification. Several assays have been developed to detect FMDV that use 5’-nuclease assay (TaqMan®) system to detect PCR amplicons (Callahan et al. 2002; Oem et al. 2005; Reid et al. 2002). Other formats exploited for FMDV-specific rRT-PCR assays include the use of modified minor groove binder (MGB) probes (McKillen et al. 2011; Moniwa et al. 2007), hybridisation probes (Moonen et al. 2003), Primer-probe energy transfer (PriProET: Rasmussen et al. 2003) and RT-linear-after-the-exponential PCR (LATE PCR: Reid et al. 2010). In order to minimise human operator errors and increase assay throughput, these assays can be automated using robots for nucleic acid extraction (Reid et al.; Moonen et al. 2003). Together with the implementation of quality control systems, these improvements have increased the acceptance of the rRT-PCR assays for routine diagnostic purposes. In addition to use in centralised laboratories, there are opportunities to deploy rRT-PCR technologies close to the animals with suspect clinical signs. These test formats may be particularly suitable for use in FMD-endemic areas such as countries within sub-Saharan Africa where the time taken to collect and dispatch samples to a laboratory for disease investigation can be protracted. Work in this area has explored the use of new hardware platforms to allow PCR testing to be deployed into the field for use by non-specialists (Callahan et al. 2002; Hearps, Zhang & Alexandersen 2002; King et al. 2008). The focus of current work is the development of hardware platforms (Madi et al. 2011) incorporating a simple-to-use and robust template extraction process such that all the steps of the assay can be performed without user intervention. These steps include (1) nucleic acid extraction, (2) PCR set-up, (3) amplification and (4) unambiguous calling of results. The use of homogeneous systems has previously been recognised an important aspect for the implementation of molecular methods for field detection of FMDV (Hearps et al. 2002). Currently, there is only limited access to these technologies outside of Europe and North America. In addition to the performance of these equipment and assays, the availability and cost of consumables, as well as mechanisms to locally service the machines (in the event of equipment failure) will be important factors for the routine use of these tests in countries within sub-Saharan Africa.
The recent development of portable equipment for PCR has made molecular diagnosis of FMD in the field an achievable goal. However, this approach relies on precision thermocycling requiring instrumentation which can be fragile, prohibitively expensive and that will require decontamination when transferred from one site to another. As an alternative to PCR, isothermal (single temperature) amplification methods for the detection of FMDV have been developed. Since the specific amplification step for both nucleic acid sequence based amplification (NASBA) and reverse-transcription loop-mediated amplification (RT-LAMP) formats occurs at a constant temperature, there is less reliance upon expensive equipment and there is obvious potential to use of these assays as the basis of an inexpensive (or even disposable) molecular test. Two different isothermal strategies: NASBA and RT-LAMP are discussed in this review. Nucleic acid sequence based amplification technology is a continuous, isothermal and enzyme-based method to amplify single stranded RNA and is therefore particularly suited to the detection of viruses with RNA genomes such as FMDV (Compton 1991). Assays employ three enzymes: T7 RNA polymerase, reverse-transcriptase and ribonuclease-H, a set of target-specific forward and reverse oligonucleotide primers and two types of detection probes. The forward primer has a 5’ extension containing the promoter sequence for T7 bacteriophage DNA-dependent RNA polymerase, while the reverse primer has a 5’ extension containing a complementary binding sequence for a DNA oligonucleotide detection probe. Using an FMDV specific NASBA assay (Collins et al. 2002), concordant results between NASBA and rRT-PCR were generated for 87% of FMD samples (Lau et al. 2008). Simple enzyme-linked oligonucleotide capture NASBA formats using ELISA plate readers have also been evaluated and these show promise for use in laboratories where more expensive equipment may not be available (Lau et al. 2008). Reverse-transcription loop-mediated amplification is an isothermal autocyling strand-displacement DNA synthesis technique which utilises four specific primers to recognise six regions of the target genome (Notomi et al. 2000). Formation of loop structures enables explosive polymerase-based enzymatic amplification, which generates double-stranded, multi-sized amplicons. Pan-serotypic RT-LAMP assays have been designed for FMDV (Dukes, King & Alexandersen 2006; Li et al. 2009). Validation data indicates that RT-LAMP has equivalent analytical sensitivity to rRT-PCR and may be less sensitive to inhibition by problematic sample matrices such as OP fluids and faecal samples. Reverse-transcription loop-mediated amplification products are generated in abundance and can be detected using equipment to monitor turbidity, agarose gels or real-time PCR machines. Furthermore, it is also possible to visualise dual-labelled LAMP amplicons using novel lateral flow devices (James et al. 2010). In addition, changes in free-cation (Mg2+) concentration in positive RT-LAMP reactions can be visualised using a colour change of a dye indicator such as hydroxynaphthol blue (Bearinger et al. 2011). Together with simple methods to prepare template RNA, these simple readouts for RT-LAMP may prove useful in field settings.
|
Using genome sequence data to define trans-boundary movements of foot-and-mouth disease virus
|
|
Once FMDV has been detected, nucleotide sequencing is a useful tool used to compare field strains with reference viruses and allows important phenotypic characteristics to be elucidated, such as antigenic determinants present on the viral capsid. Genetic characterisation of foot-and-mouth disease virus routinely uses VP1 sequence data generated by RT-PCR. This region of the FMDV genome is approximately 630 nucleotides in length and encodes a protein (1D) that comprises an important component of the FMD viral capsid. These sequences are used to categorise field strains into discrete sub-groups (or topotypes) which frequently show geographical clustering based on the historical distribution of the virus (Di Nardo, Knowles & Paton 2011). These analyses provide evidence for the transboundary movements of FMDV and provide critical support to regional and country-level programmes to control FMD. Within sub-Saharan Africa, three main epidemiological pools of FMDV have been recognised (Paton, Sumption & Charleston 2009): West Africa comprising serotypes O, A, SAT 1 and SAT 2; East Africa comprising serotypes O, A, SAT 1, SAT 2 and SAT 3 and Southern Africa with serotypes SAT 1, SAT 2 and SAT 3. Recent FMD outbreaks in the United Kingdom (during 2001 and 2007) have highlighted the limitation to which VP1 sequence data can be used to discriminate sequences from field cases of disease. VP1 is relatively short (~8% of the genome length) and as a consequence, phylogenetic trees generated from viral sequences recovered within outbreaks clusters are typically flat with poor resolution. Furthermore, limiting sequence analysis to only VP1 reduces the ability to identify broad-scale recombination events that may drive step-changes in the generation of new genetic and antigenic variants. Identifying the sources of FMD outbreaks can play an important role in disease control: however, this can be confounded by incomplete epidemiological evidence and the numerous routes by which the virus can spread (via aerosols, movements of infected animals or their products, and spread of fomites on contaminated persons and objects). Recent advances in laboratory methodologies allow rapid sequencing of complete FMDV genomes (Abdul-Hamid et al. 2011; Cottam et al. 2009a). For the first time, this opens up the potential for using genome sequencing to reconstruct virus transmission trees with extremely high resolution and to reveal and identify the origin of unresolved transmission events within discrete infection clusters quickly. Using full FMDV genome sequences determined from field samples collected from the 2001 FMD outbreak in the UK, it was shown that transmission events at the level of farm-to-farm spread could be reconstructed (Cottam et al. 2006, 2008b). Known patterns of spread of the virus were be reproduced by statistical parsimony-based analyses of this data. However, in some cases, these genetic data supported transmission histories different from those suggested by conventional contact tracing studies (Cottam et al. 2006). During the 2007 FMD outbreak in the UK, full genome sequencing was used in real-time to support epidemiological investigations (Cottam et al. 2008a). Analyses of these data have revealed the most likely chain of transmission events, and predicted undisclosed infected premises prior to their discovery by serological surveillance. Thus, for the first time, results indicate that full-genome sequencing can be used for fine-scale epidemiology to reveal and identify the origin of FMDV causing outbreaks.
|
Foot-and-mouth disease viral evolutionary dynamics
|
|
Foot-and-mouth disease evolves rapidly due to its large population size, high replication rate and poor proof-reading ability of its RNA-dependent RNA polymerase. Within cells, FMDV exists as heterogeneous populations comprising similar but non-identical genomes. Consensus (Sanger) sequencing identifies the predominant sequence present in a sample, but does not provide any information regarding the structure of minority variants that are present. Next-Generation Sequencing (NGS) techniques offer an unprecedented ‘step-change’ increase in the amount of sequence data that can be generated from a sample to reveal nucleotide substitutions present in only a small fraction of the population. Using NGS performed on a Genome Analyzer platform (Illumina), the viral populations within bovine epithelial samples (foot lesions) have been determined (Wright et al. 2011). This approach revealed the fine polymorphic sub-structure of the viral population, from nucleotide variants present at just below 50% frequency to those present at fractions of 1%. Some of the higher frequency polymorphisms identified encoded changes in the heparan sulphate binding site revealing intermediate stages in the evolution of a tissue-culture adapted viral genome upon replication within a mammalian host. Conventional Sanger sequencing and NGS methods can provide powerful datasets that have the potential to revolutionise our understanding of the patterns of viral evolution and factors that impact upon FMDV circulation in endemic countries. Capital equipment and running cost to undertake sequencing studies are decreasing all the time. Regional and national centres of molecular and sequencing expertise are now becoming established in some African countries to provide support for research into the epidemiology of livestock diseases such as FMD. Although currently limited to use in specialised molecular facilities, there is a prospect that new more affordable formats of NGS technologies will become available soon.
|
Diagnostic challenges for sub-Saharan Africa
|
|
This short review describes new tools that have recently been developed to support the diagnosis and epidemiology of FMD. The driving force for these improvements has largely been influenced by the priorities of developed countries in Europe and North America with FMD-free (without vaccination) status. However, it is important to recognise that these technologies also show considerable promise for use in FMD-endemic countries, although further modifications may be required to tailor these tests for use in these regions. The presence of multiple serotypes of FMD in the majority of sub-Saharan African countries can influence local disease diagnostic priorities. Rather than pan-serotypic FMDV assays that are developed in FMD-free countries, a particular priority for endemic countries is the development of serotype-specific (and lineage-specific) typing assays that can be used rapidly to monitor incursions of exotic and
emerging FMDV lineages into new geographical regions. The high genetic diversity of circulating FMDV strains (particularly SAT serotypes) provides a constant challenge for diagnostic laboratories in FMD endemic countries in Africa, and it is important that on-going validation is undertaken to ensure tests remain fit for purpose and are able to detect new viruses as they continue to evolve. Furthermore, it is critical that robust protocols for sample collection and archiving in the field are adopted (Belsham et al. 2011). Our understanding of FMD epidemiology in Africa is patchy and currently suffers from a lack of good quality laboratory and sequence data. The global control of FMD strategy currently being developed jointly by OIE and FAO is expected to rely heavily on the use of these next generation tools for more accurate and rapid diagnosis of FMD at the local level. However, without improvements to the quality and coverage of samples collected from field outbreaks, it is unlikely that the epidemiological picture of the disease in the region will become clear enough to make a significant impact upon disease control.
These new diagnostic tools can play a critical role in our ability to detect and monitor the spread of FMD in endemic regions of the world. However, it is also important to recognise that effective monitoring and control of FMD is reliant upon adequate resources, these are principally financial but also include availability of trained field personnel and a strong supporting laboratory infrastructure. In addition, international cooperation, transparency between different countries, sharing of epidemiological data and ownership of
disease are also key factors in the control of important trans-boundary disease such as FMD.
The authors of this article are supported by grant funding from UK Department of Environment, Food and Rural Affairs (Defra SE1124, SE1126, SE2938 and SE2937), EU NoE EPIZONE (Contract No. FOOD-CT-2006-016236) and the Combating Infectious Diseases of Livestock for International Development (CIDLID) project funded by DFiD/BBSRC/Scottish Government (Towards the strategic control of foot-and-mouth disease in Africa: New techniques for a neglected problem). The authors also acknowledge Southern African Centre for Infectious Disease Surveillance (SACIDS) for providing
travel support (to DK) to present this paper at the 1st One Health Conference in Africa, Johannesburg, July 2011.
Abdul-Hamid, N.F., Fırat-Saraç, M., Radford, A.D., Knowles, N.J. & King, D.P., 2011, ‘Comparative sequence analysis of representative foot-and-mouth disease virus genomes from Southeast Asia’, Virus Genes 43, 41–45.
http://dx.doi.org/10.1007/s11262-011-0599-3,
PMid:21479678
Alexandersen, S., Quan, M., Murphy, C., Knight, J. & Zhang, Z., 2003, ‘Studies of quantitative parameters of virus excretion and transmission in pigs and cattle experimentally infected with foot-and-mouth disease virus’, Journal of Comparative Pathology 129, 268–282.
http://dx.doi.org/10.1016/S0021-9975(03)00045-8 org/10.1371/journal.pone.0014621 Bearinger, J.P., Dugan, L.C., Baker, B.R., Hall, S.B., Ebert, K., Mioulet, V. et al., 2011, ‘Development
and initial results of a low cost, disposable, point-of-care testing device for pathogen detection’, IEEE Transactions
on Biomedical Engineering 58, 805–808.
http://dx.doi.org/10.1109/TBME.2010.2089054 org/10.1371/journal.pone.0014621,
PMid:21342806
Belsham, G.J., Jamal, S.M., Tjørnehøj, K. & Bøtner, A., 2011, ‘Rescue of foot-and-mouth disease viruses that are pathogenic for cattle from preserved viral RNA samples’, PLoS One 6, e14621.
http://dx.doi.org/10.1371/journal.pone.0014621,
PMid:21298025
Brehm, K.E., Ferris, N.P., Lenk, M., Riebe, R. & Haas, B., 2009, ‘Highly sensitive fetal goat tongue cell line for detection and isolation of foot-and-mouth disease virus’, Journal of Clinical Microbiology 47, 3156–3160.
http://dx.doi.org/10.1128/JCM.00510-09,
PMid:19656987
Brown, F., 2003, ‘The history of research in foot-and-mouth disease’, Virus Research 91, 3–7.
http://dx.doi.org/10.1016/S0168-1702(02)00268-X Callahan, J.D., Brown, F., Osorio, F.A., Sur, J.H., Kramer, E., Long, G.W. et al., 2002, ‘Use
of a portable real-time reverse transcriptase-polymerase chain reaction assay for rapid detection of foot-and-mouth
disease virus’, Journal of the American Medical Association 220, 1636–1642. Collins, R.A., Ko, L.S., Fung, K.Y., Lau, L.T., Xing, J. & Yu, A.C.H., 2002b, ‘A method to detect major serotypes of foot-and-mouth disease virus’, Biochemistry Biophysics Research Communication 297, 267–274.
http://dx.doi.org/10.1016/S0006-291X(02)02178-2 Compton, J., 1991, ‘Nucleic acid sequence-based amplification’, Nature 350, 91–92.
http://dx.doi.org/10.1038/350091a0,
PMid:1706072
Cottam, E.M., Haydon, D.T., Paton, D.J., Gloster, J., Wilesmith, J.W., Ferris, N.P. et al., 2006, ‘Molecular epidemiology of the foot-and-mouth disease virus outbreak in the United Kingdom in 2001’, Journal of Virology 80, 11274–11282.
http://dx.doi.org/10.1128/JVI.01236-06,
PMid:16971422
Cottam, E.M., Wadsworth, J., Shaw, A.E., Rowlands, R.J., Goatley, L., Maan, S. et al., 2008a, ‘Transmission pathways of foot-and-mouth disease virus in the United Kingdom in 2007’, PloS Pathogens 4, e1000050.
http://dx.doi.org/10.1371/journal.ppat.1000050,
PMid:18421380
Cottam, E.M., Thébaud, G., Wadsworth, J., Gloster, J., Mansley, L., Paton, D.J. et al., 2008b, ‘Integrating genetic and epidemiological data to determine transmission pathways of foot-and-mouth disease virus’, Proceedings of the Royal Society B 275, 887–895.
http://dx.doi.org/10.1098/rspb.2007.1442,
PMid:18230598
Cottam, E.M., Wadsworth, J., Knowles, N.J. & King, D.P., 2009a, ‘Full sequencing of viral genomes: practical strategies used for the amplification and characterization of foot-and-mouth disease virus’, Methods in Molecular Biology 551, 217–230.
http://dx.doi.org/10.1007/978-1-60327-999-4_17,
PMid:19521878
De Castro, M.P., 1964, ‘Behaviour of the foot-and-mouth disease virus in cell cultures; susceptibility of the IB-RS-2 cell line’, Archivos do Instituto Biologico, São Paulo 31, 63–78. Di Nardo, A., Knowles, N.J. & Paton, D.J., 2011, ‘Combining livestock trade patterns
with phylogenetics to help understand the spread of foot-and-mouth disease virus in sub-Saharan Africa,
the middle East and Southeast Asia’, Scientific and Technical Review of the OIE 30, 63–85. Dukes, J.P., King, D.P. & Alexandersen, S., 2006, ‘Novel reverse transcription loop-mediated isothermal amplification for rapid detection of foot-and-mouth disease virus’, Archives of Virology 151, 1093–1106.
http://dx.doi.org/10.1007/s00705-005-0708-5,
PMid:16453084
Ferris, N.P. & Dawson, M., 1988, ‘Routine application of enzyme-linked immunosorbent assay in comparison with complement fixation for the diagnosis of foot-and-mouth and swine vesicular diseases’, Veterinary Microbiology 16, 201–209.
http://dx.doi.org/10.1016/0378-1135(88)90024-7 Ferris, N.P., Hutchings, G.H., Reid, S.M., King, D.P., Ebert, K., Paton, D.J. et al., M., 2009, ‘Development and laboratory validation of a lateral flow device for the detection of foot-and-mouth disease virus in clinical samples’, Journal of Virological Methods 155, 10–17.
http://dx.doi.org/10.1016/j.jviromet.2008.09.009,
PMid:18848845
Ferris, N.P., Nordengrahn, A., Hutchings, G.H., Paton, D.J., Kristersson, T., Brocchi, E. et al., 2010, ‘Development and laboratory validation of a lateral flow device for the detection of serotype SAT 2 foot-and-mouth disease viruses in clinical samples’, Journal of Virological Methods 163, 474–476.
http://dx.doi.org/10.1016/j.jviromet.2009.09.022,
PMid:19815031
Ferris, N.P., Grazioli, S., Hutchings, G.H. & Brocchi, E., 2011, ‘Validation of a recombinant integrin αvβ6/monoclonal
antibody based antigen ELISA for the diagnosis of foot-and-mouth disease’, Journal of Virological Methods 175, 253–260.
http://dx.doi.org/10.1016/j.jviromet.2011.05.026,
PMid:21635921
Hearps, A., Zhang, Z. & Alexandersen, S., 2002, ‘Evaluation of a portable Cepheid SmartCycler real-time PCR machine for the rapid diagnosis of foot-and-mouth disease’, Veterinary Record 150, 625–628.
http://dx.doi.org/10.1136/vr.150.20.625,
PMid:12046786
James, H.E., Ebert, K., McGonigle, R., Reid, S.M., Boonham, N., Tomlinson, J.A. et al., 2010, ‘Detection of African swine fever virus by loop-mediated isothermal amplification’, Journal of Virological Methods 164, 68–74.
http://dx.doi.org/10.1016/j.jviromet.2009.11.034,
PMid:19963011
King, D.P., Dukes, J.P., Reid, S.M., Ebert, K., Shaw, A.E., Mills, C.E. et al., 2008, ‘Prospects for rapid diagnosis of foot-and-mouth disease in the field using reverse transcriptase-PCR’, Veterinary Record 162, 315–316.
http://dx.doi.org/10.1136/vr.162.10.315,
PMid:18326844
Lau, L.T., Reid, S.M., King, D.P., Lau, A.M., Shaw, A.E., Ferris, N.P. et al., 2008, ‘Detection of foot-and-mouth disease virus by nucleic acid sequence-based amplification (NASBA)’, Veterinary Microbiology 126, 101–110.
http://dx.doi.org/10.1016/j.vetmic.2007.07.008,
PMid:17728080
Li, J., Chen, Q., Xiong, W. & Fang, X.E., 2009, ‘Establishment of RT-LAMP for rapid detection of
foot-and-mouth disease virus’, Bing Du Xue Bao 25, 137–142.
PMid:19678569
Madi, M., Hamilton, A., Squirrell, D., Mioulet, V., Evans, P., Lee, M. et al., 2011, ‘Rapid detection of
foot-and-mouth disease virus using a field-portable nucleic acid extraction and real-time PCR amplification platform’,
Veterinary Journal (submitted).
PMid:21668486
McKillen, J., McMenamy, M., Reid, S.M., Duffy, C., Hiertner, B., King, D.P. et al., 2011, ‘Pan-serotypic detection of foot-and-mouth disease virus using a minor groove binder probe reverse transcription polymerase chain reaction assay’, Journal of Virological Methods 174, 117–119.
http://dx.doi.org/10.1016/j.jviromet.2011.03.008,
PMid:21419170
Meyer, R.F., Brown, C.C., House, C., House, J.A. & Molitor, T.W., 1991, ‘Rapid and sensitive detection of foot-and-mouth disease virus in tissues by enzymatic RNA amplification of the polymerase gene’, Journal of Virological Methods 34, 161–172.
http://dx.doi.org/10.1016/0166-0934(91)90096-I Moonen, P., Boonstra, J., Van der Honing, R.H., Leendertse, C.B., Jacob, L. & Dekker, A., 2003, ‘Validation of a light Cycler-based reverse transcriptase polymerase chain reaction for the detection of foot-and-mouth disease virus’, Journal of Virological Methods 113, 35–41.
http://dx.doi.org/10.1016/S0166-0934(03)00220-9 Moniwa, M., Clavijo, A., Li, M., Collignon, B. & Kitching, P.R., 2007, ‘Performance of a foot-and-disease virus reverse transcription-polymerase chain reaction with amplification controls between three real-time instruments’, Journal of Veterinary Diagnostic Investigation 19, 9–20.
http://dx.doi.org/10.1177/104063870701900103,
PMid:17459827
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N. et al., 2000, ‘Loop-mediated isothermal amplification of DNA’, Nucleic Acids Research 28, E63.
http://dx.doi.org/10.1093/nar/28.12.e63 Oem, J.K., Kye, S.J., Lee, K.N., Kim, J.Y., Park, J.H., Joo, Y.S. et al., 2005, ‘Development of a
light cycler-based reverse transcriptase polymerase reaction for the detection of foot-and-mouth disease virus’,
Journal of Veterinary Science 6, 207–212.
PMid:16131823
Paton, D.J., Sumption, K.J. & Charleston, B., 2009, ‘Options for control of foot-and-mouth disease: knowledge, capability and policy’, Philosophical Transactions of the Royal Society London B Biology Science 364, 2657–2567.
http://dx.doi.org/10.1098/rstb.2009.0100,
PMid:19687036
Rasmussen, T.B., Uttenthal, A., De Stricker, K., Belak, S. & Storgaard, T., 2003, ‘Development of a novel quantitative real-time RT-PCR assay for the simultaneous detection of all serotypes of foot-and-mouth disease virus’, Archives of Virology 148, 2005–2021.
http://dx.doi.org/10.1007/s00705-003-0145-2,
PMid:14551821
Reid, S.M., Ferris, N.P., Hutchings, G.H., Samuel, A.R. & Knowles, N.J., 2000, ‘Primary diagnosis of foot-and-mouth disease by reverse transcription polymerase chain reaction’, Journal of Virological Methods 89, 167–176.
http://dx.doi.org/10.1016/S0166-0934(00)00213-5 Reid, S.M., Ferris, N.P., Hutchings, G.H., Zhang, Z., Belsham, G.J. & Alexandersen, S., 2002, ‘Detection of all seven serotypes of foot-and-mouth disease virus by real-time, fluorogenic reverse transcription polymerase chain reaction assay’, Journal of Virological Methods 105, 67–80.
http://dx.doi.org/10.1016/S0166-0934(02)00081-2 Reid, S.M., Grierson, S.S., Ferris, N.P., Hutchings, G.H. & Alexandersen, S., 2003, ‘Evaluation of automated RT-PCR to accelerate the laboratory diagnosis of foot-and-mouth disease virus’, Journal of Virological Methods 107, 129–139.
http://dx.doi.org/10.1016/S0166-0934(02)00210-0 Reid, S.M., Pierce, K.E., Mistry, R., Bharya, S., Dukes, J.P., Volpe, C. et al., 2010, ‘Pan-serotypic detection of foot-and-mouth disease virus by RT linear-after-the exponential PCR’, Molecular and Cellular Probes 24, 250–255.
http://dx.doi.org/10.1016/j.mcp.2010.04.004,
PMid:20433917
Rodríguez, A., Núñez, J.I., Nolasco, G., Ponz, F., Sobrino, F. & De Blas, C., 1994, ‘Direct PCR detection of foot-and-mouth disease virus’, Journal of Virological Methods 47, 345–349.
http://dx.doi.org/10.1016/0166-0934(94)90030-2
Snowdon, W.A., 1966, ‘Growth of foot-and mouth disease virus in monolayer cultures of calf thyroid cells’, Nature 210, 1079–1080.
http://dx.doi.org/10.1038/2101079a0,
PMid:4288087
Wright, C.F., Morelli, M.J., Thébaud, G., Knowles, N.J., Herzyk, P., Paton, D.J. et al., 2011, ‘Beyond
the consensus: dissecting within-host population diversity of foot-and-mouth disease virus using next-generation sequencing’,
Journal of Virology 85, 2266–2275.
http://dx.doi.org/10.1128/JVI.01396-10,
PMid:21159860
|